Lewinella sp. W8
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Saprospiria; Saprospirales; Lewinellaceae; Lewinella; unclassified Lewinella
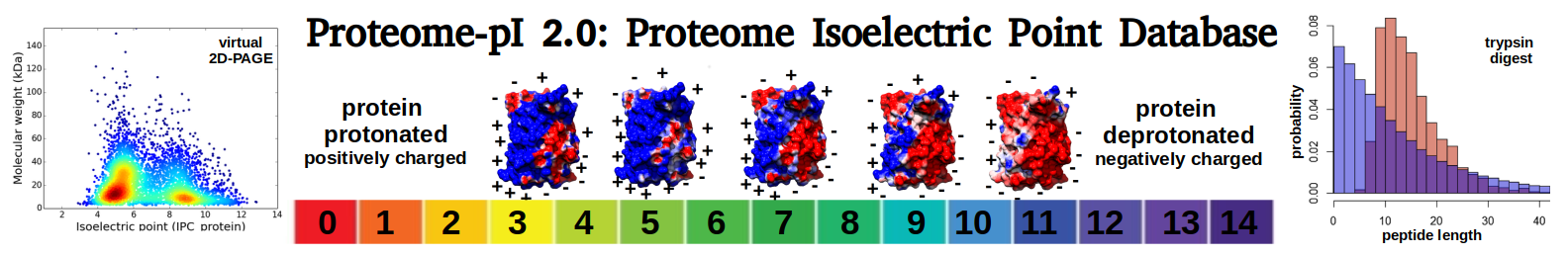
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 4697 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions



Protein with the lowest isoelectric point:
>tr|A0A7H5ETJ1|A0A7H5ETJ1_9BACT CHAT domain-containing protein OS=Lewinella sp. W8 OX=2528208 GN=E1J53_0004175 PE=4 SV=1
MM1 pKa = 7.62NYY3 pKa = 10.03LHH5 pKa = 7.03SLRR8 pKa = 11.84LLSGLFFVVLAFGGLSAQVTQDD30 pKa = 3.63FSTSTPGSHH39 pKa = 6.95HH40 pKa = 6.39YY41 pKa = 10.57VDD43 pKa = 4.25PLSPFTEE50 pKa = 5.36HH51 pKa = 7.31ILPATMAPQTQPNAATMGAIPGYY74 pKa = 9.57QVTYY78 pKa = 9.79TPTRR82 pKa = 11.84DD83 pKa = 3.11RR84 pKa = 11.84DD85 pKa = 3.69AGGGLTDD92 pKa = 4.0GEE94 pKa = 4.5LFGVVDD100 pKa = 3.82YY101 pKa = 10.0TVAPTPDD108 pKa = 3.41NDD110 pKa = 3.85MGSLTIDD117 pKa = 3.34IAANPPPSGNNNIYY131 pKa = 10.45VLQDD135 pKa = 3.38PDD137 pKa = 3.5GLATLRR143 pKa = 11.84FNPVVVDD150 pKa = 4.15GSTMFSMSYY159 pKa = 9.86IVANTSYY166 pKa = 11.14EE167 pKa = 4.17NGDD170 pKa = 3.63GADD173 pKa = 4.06DD174 pKa = 4.5RR175 pKa = 11.84IEE177 pKa = 4.24IYY179 pKa = 10.7LINTATGDD187 pKa = 3.3RR188 pKa = 11.84TMLFSAINEE197 pKa = 4.74AIPTTEE203 pKa = 3.25VWTTLTADD211 pKa = 3.93LSGLAGATVQLVIEE225 pKa = 4.83FDD227 pKa = 3.9TNASAEE233 pKa = 3.98EE234 pKa = 3.83MAIDD238 pKa = 4.56NIIFSTGVVEE248 pKa = 5.55ADD250 pKa = 2.81WPEE253 pKa = 4.04CTEE256 pKa = 4.22PSLAALGQSPATVCPGVPFRR276 pKa = 11.84MEE278 pKa = 3.46ISGSLGNATEE288 pKa = 4.05WVIYY292 pKa = 10.48SDD294 pKa = 4.65AEE296 pKa = 4.31GTQEE300 pKa = 4.14VARR303 pKa = 11.84TSSDD307 pKa = 3.07QYY309 pKa = 11.67DD310 pKa = 3.75FFGGAEE316 pKa = 4.41LGTTYY321 pKa = 10.68YY322 pKa = 10.84VRR324 pKa = 11.84GEE326 pKa = 4.28GGCASSMDD334 pKa = 4.0LTALALTIDD343 pKa = 5.39DD344 pKa = 5.1GDD346 pKa = 4.63CEE348 pKa = 4.48PNAPPSTSFEE358 pKa = 4.17DD359 pKa = 3.66ALASQEE365 pKa = 4.82DD366 pKa = 4.59YY367 pKa = 11.63VDD369 pKa = 3.26TGNPVIFRR377 pKa = 11.84FLEE380 pKa = 4.17NNQGQPTVTTGGRR393 pKa = 11.84GKK395 pKa = 10.39EE396 pKa = 3.91LGFITAFAPTRR407 pKa = 11.84LGTSGEE413 pKa = 4.15AGLTDD418 pKa = 4.05GDD420 pKa = 4.26ALGVTDD426 pKa = 5.86DD427 pKa = 4.46PDD429 pKa = 3.49AAFTDD434 pKa = 4.54GEE436 pKa = 4.38KK437 pKa = 11.18GFVFEE442 pKa = 4.88DD443 pKa = 3.51TDD445 pKa = 4.14GLVSAVFAPIDD456 pKa = 3.43IAGLSGVTVSFDD468 pKa = 3.13YY469 pKa = 10.58FIRR472 pKa = 11.84STSYY476 pKa = 10.08EE477 pKa = 4.09SSSTGTDD484 pKa = 2.36RR485 pKa = 11.84FTAVILNEE493 pKa = 4.24SFGLLEE499 pKa = 4.93EE500 pKa = 4.79IFDD503 pKa = 4.13AAADD507 pKa = 4.19GSGGFPDD514 pKa = 4.07LTIGQWTTVTYY525 pKa = 9.63TITEE529 pKa = 4.34SYY531 pKa = 10.71DD532 pKa = 3.16DD533 pKa = 5.24PIRR536 pKa = 11.84LTIQADD542 pKa = 3.62LDD544 pKa = 4.08AGSEE548 pKa = 4.19RR549 pKa = 11.84IFIDD553 pKa = 3.18NVSFSAGTVVCQDD566 pKa = 2.85EE567 pKa = 4.63AAPVIACPEE576 pKa = 4.87PISVDD581 pKa = 3.36VGADD585 pKa = 3.35CMATAEE591 pKa = 4.32FTATATDD598 pKa = 4.02DD599 pKa = 3.71CSSTVTITYY608 pKa = 10.12SQDD611 pKa = 2.79SGTNFPVGVTTVTATATDD629 pKa = 3.75EE630 pKa = 4.32TGKK633 pKa = 10.88SSTCTFDD640 pKa = 3.18VTVNDD645 pKa = 3.56VTAPTLDD652 pKa = 3.84CPEE655 pKa = 4.11VTVKK659 pKa = 10.73LDD661 pKa = 3.4STFSCDD667 pKa = 4.06FVVISNTDD675 pKa = 3.93FDD677 pKa = 6.04PIAADD682 pKa = 3.16NCEE685 pKa = 3.76GVTLLSANGLSSLVGDD701 pKa = 4.53TIFKK705 pKa = 7.96EE706 pKa = 4.57TTEE709 pKa = 4.29LTWTATDD716 pKa = 3.59ASGLTATCTQQINVIVPPSCFPTSVTEE743 pKa = 3.93QDD745 pKa = 3.36PSDD748 pKa = 3.78FANVVAIPNPFRR760 pKa = 11.84EE761 pKa = 3.94AVTINFEE768 pKa = 4.49LPFMDD773 pKa = 3.93EE774 pKa = 3.87VGIDD778 pKa = 3.57VLDD781 pKa = 3.45VSGRR785 pKa = 11.84VVQSARR791 pKa = 11.84VNPNGDD797 pKa = 2.95RR798 pKa = 11.84TYY800 pKa = 10.8SWRR803 pKa = 11.84WDD805 pKa = 3.38ATDD808 pKa = 3.35AAGGRR813 pKa = 11.84AAAGMYY819 pKa = 8.96FVRR822 pKa = 11.84LRR824 pKa = 11.84ARR826 pKa = 11.84GQLYY830 pKa = 7.84TKK832 pKa = 10.38RR833 pKa = 11.84IVLTRR838 pKa = 3.63
MM1 pKa = 7.62NYY3 pKa = 10.03LHH5 pKa = 7.03SLRR8 pKa = 11.84LLSGLFFVVLAFGGLSAQVTQDD30 pKa = 3.63FSTSTPGSHH39 pKa = 6.95HH40 pKa = 6.39YY41 pKa = 10.57VDD43 pKa = 4.25PLSPFTEE50 pKa = 5.36HH51 pKa = 7.31ILPATMAPQTQPNAATMGAIPGYY74 pKa = 9.57QVTYY78 pKa = 9.79TPTRR82 pKa = 11.84DD83 pKa = 3.11RR84 pKa = 11.84DD85 pKa = 3.69AGGGLTDD92 pKa = 4.0GEE94 pKa = 4.5LFGVVDD100 pKa = 3.82YY101 pKa = 10.0TVAPTPDD108 pKa = 3.41NDD110 pKa = 3.85MGSLTIDD117 pKa = 3.34IAANPPPSGNNNIYY131 pKa = 10.45VLQDD135 pKa = 3.38PDD137 pKa = 3.5GLATLRR143 pKa = 11.84FNPVVVDD150 pKa = 4.15GSTMFSMSYY159 pKa = 9.86IVANTSYY166 pKa = 11.14EE167 pKa = 4.17NGDD170 pKa = 3.63GADD173 pKa = 4.06DD174 pKa = 4.5RR175 pKa = 11.84IEE177 pKa = 4.24IYY179 pKa = 10.7LINTATGDD187 pKa = 3.3RR188 pKa = 11.84TMLFSAINEE197 pKa = 4.74AIPTTEE203 pKa = 3.25VWTTLTADD211 pKa = 3.93LSGLAGATVQLVIEE225 pKa = 4.83FDD227 pKa = 3.9TNASAEE233 pKa = 3.98EE234 pKa = 3.83MAIDD238 pKa = 4.56NIIFSTGVVEE248 pKa = 5.55ADD250 pKa = 2.81WPEE253 pKa = 4.04CTEE256 pKa = 4.22PSLAALGQSPATVCPGVPFRR276 pKa = 11.84MEE278 pKa = 3.46ISGSLGNATEE288 pKa = 4.05WVIYY292 pKa = 10.48SDD294 pKa = 4.65AEE296 pKa = 4.31GTQEE300 pKa = 4.14VARR303 pKa = 11.84TSSDD307 pKa = 3.07QYY309 pKa = 11.67DD310 pKa = 3.75FFGGAEE316 pKa = 4.41LGTTYY321 pKa = 10.68YY322 pKa = 10.84VRR324 pKa = 11.84GEE326 pKa = 4.28GGCASSMDD334 pKa = 4.0LTALALTIDD343 pKa = 5.39DD344 pKa = 5.1GDD346 pKa = 4.63CEE348 pKa = 4.48PNAPPSTSFEE358 pKa = 4.17DD359 pKa = 3.66ALASQEE365 pKa = 4.82DD366 pKa = 4.59YY367 pKa = 11.63VDD369 pKa = 3.26TGNPVIFRR377 pKa = 11.84FLEE380 pKa = 4.17NNQGQPTVTTGGRR393 pKa = 11.84GKK395 pKa = 10.39EE396 pKa = 3.91LGFITAFAPTRR407 pKa = 11.84LGTSGEE413 pKa = 4.15AGLTDD418 pKa = 4.05GDD420 pKa = 4.26ALGVTDD426 pKa = 5.86DD427 pKa = 4.46PDD429 pKa = 3.49AAFTDD434 pKa = 4.54GEE436 pKa = 4.38KK437 pKa = 11.18GFVFEE442 pKa = 4.88DD443 pKa = 3.51TDD445 pKa = 4.14GLVSAVFAPIDD456 pKa = 3.43IAGLSGVTVSFDD468 pKa = 3.13YY469 pKa = 10.58FIRR472 pKa = 11.84STSYY476 pKa = 10.08EE477 pKa = 4.09SSSTGTDD484 pKa = 2.36RR485 pKa = 11.84FTAVILNEE493 pKa = 4.24SFGLLEE499 pKa = 4.93EE500 pKa = 4.79IFDD503 pKa = 4.13AAADD507 pKa = 4.19GSGGFPDD514 pKa = 4.07LTIGQWTTVTYY525 pKa = 9.63TITEE529 pKa = 4.34SYY531 pKa = 10.71DD532 pKa = 3.16DD533 pKa = 5.24PIRR536 pKa = 11.84LTIQADD542 pKa = 3.62LDD544 pKa = 4.08AGSEE548 pKa = 4.19RR549 pKa = 11.84IFIDD553 pKa = 3.18NVSFSAGTVVCQDD566 pKa = 2.85EE567 pKa = 4.63AAPVIACPEE576 pKa = 4.87PISVDD581 pKa = 3.36VGADD585 pKa = 3.35CMATAEE591 pKa = 4.32FTATATDD598 pKa = 4.02DD599 pKa = 3.71CSSTVTITYY608 pKa = 10.12SQDD611 pKa = 2.79SGTNFPVGVTTVTATATDD629 pKa = 3.75EE630 pKa = 4.32TGKK633 pKa = 10.88SSTCTFDD640 pKa = 3.18VTVNDD645 pKa = 3.56VTAPTLDD652 pKa = 3.84CPEE655 pKa = 4.11VTVKK659 pKa = 10.73LDD661 pKa = 3.4STFSCDD667 pKa = 4.06FVVISNTDD675 pKa = 3.93FDD677 pKa = 6.04PIAADD682 pKa = 3.16NCEE685 pKa = 3.76GVTLLSANGLSSLVGDD701 pKa = 4.53TIFKK705 pKa = 7.96EE706 pKa = 4.57TTEE709 pKa = 4.29LTWTATDD716 pKa = 3.59ASGLTATCTQQINVIVPPSCFPTSVTEE743 pKa = 3.93QDD745 pKa = 3.36PSDD748 pKa = 3.78FANVVAIPNPFRR760 pKa = 11.84EE761 pKa = 3.94AVTINFEE768 pKa = 4.49LPFMDD773 pKa = 3.93EE774 pKa = 3.87VGIDD778 pKa = 3.57VLDD781 pKa = 3.45VSGRR785 pKa = 11.84VVQSARR791 pKa = 11.84VNPNGDD797 pKa = 2.95RR798 pKa = 11.84TYY800 pKa = 10.8SWRR803 pKa = 11.84WDD805 pKa = 3.38ATDD808 pKa = 3.35AAGGRR813 pKa = 11.84AAAGMYY819 pKa = 8.96FVRR822 pKa = 11.84LRR824 pKa = 11.84ARR826 pKa = 11.84GQLYY830 pKa = 7.84TKK832 pKa = 10.38RR833 pKa = 11.84IVLTRR838 pKa = 3.63
Molecular weight: 88.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7H5EYX6|A0A7H5EYX6_9BACT Glutamate--tRNA ligase OS=Lewinella sp. W8 OX=2528208 GN=gltX PE=3 SV=1
MM1 pKa = 7.59SKK3 pKa = 8.74TANKK7 pKa = 9.77IFLNVRR13 pKa = 11.84TSTWMRR19 pKa = 11.84ALTVVVMLGFGWQCYY34 pKa = 8.8HH35 pKa = 7.4LARR38 pKa = 11.84RR39 pKa = 11.84VDD41 pKa = 4.15FGEE44 pKa = 3.84LLARR48 pKa = 11.84LGSPSVWPWFVLVLAMMPLNWWLEE72 pKa = 3.84ARR74 pKa = 11.84KK75 pKa = 8.47WQLLLRR81 pKa = 11.84PFLAWPFAKK90 pKa = 9.72TLRR93 pKa = 11.84ATMVGVSLSAATPNRR108 pKa = 11.84IGEE111 pKa = 3.99IGGRR115 pKa = 11.84MLMGSRR121 pKa = 11.84EE122 pKa = 4.07EE123 pKa = 4.28AGSILTSSLLGSAAQWVAFLLLAWPALMWTAGDD156 pKa = 4.24LLADD160 pKa = 5.57RR161 pKa = 11.84IPFPVALLWPLGPGVLLLLWFGGKK185 pKa = 8.98PLLVKK190 pKa = 10.35IIRR193 pKa = 11.84WVSRR197 pKa = 11.84RR198 pKa = 11.84FSWNNAPLLEE208 pKa = 4.89GIRR211 pKa = 11.84EE212 pKa = 4.24VKK214 pKa = 10.22FGLMMRR220 pKa = 11.84AGAYY224 pKa = 9.48ACLRR228 pKa = 11.84FCVYY232 pKa = 10.48CMQLHH237 pKa = 6.47ILLGCFGLEE246 pKa = 4.03LPLLEE251 pKa = 4.51GMAGIAAIYY260 pKa = 9.19LVQAGIPLPPGLNLVTRR277 pKa = 11.84TEE279 pKa = 4.19LGLLLWNTEE288 pKa = 3.99PGAGIATVLAFGSLFAINVLLPALPAYY315 pKa = 8.73WLLVRR320 pKa = 11.84KK321 pKa = 10.19KK322 pKa = 10.26NLRR325 pKa = 3.62
MM1 pKa = 7.59SKK3 pKa = 8.74TANKK7 pKa = 9.77IFLNVRR13 pKa = 11.84TSTWMRR19 pKa = 11.84ALTVVVMLGFGWQCYY34 pKa = 8.8HH35 pKa = 7.4LARR38 pKa = 11.84RR39 pKa = 11.84VDD41 pKa = 4.15FGEE44 pKa = 3.84LLARR48 pKa = 11.84LGSPSVWPWFVLVLAMMPLNWWLEE72 pKa = 3.84ARR74 pKa = 11.84KK75 pKa = 8.47WQLLLRR81 pKa = 11.84PFLAWPFAKK90 pKa = 9.72TLRR93 pKa = 11.84ATMVGVSLSAATPNRR108 pKa = 11.84IGEE111 pKa = 3.99IGGRR115 pKa = 11.84MLMGSRR121 pKa = 11.84EE122 pKa = 4.07EE123 pKa = 4.28AGSILTSSLLGSAAQWVAFLLLAWPALMWTAGDD156 pKa = 4.24LLADD160 pKa = 5.57RR161 pKa = 11.84IPFPVALLWPLGPGVLLLLWFGGKK185 pKa = 8.98PLLVKK190 pKa = 10.35IIRR193 pKa = 11.84WVSRR197 pKa = 11.84RR198 pKa = 11.84FSWNNAPLLEE208 pKa = 4.89GIRR211 pKa = 11.84EE212 pKa = 4.24VKK214 pKa = 10.22FGLMMRR220 pKa = 11.84AGAYY224 pKa = 9.48ACLRR228 pKa = 11.84FCVYY232 pKa = 10.48CMQLHH237 pKa = 6.47ILLGCFGLEE246 pKa = 4.03LPLLEE251 pKa = 4.51GMAGIAAIYY260 pKa = 9.19LVQAGIPLPPGLNLVTRR277 pKa = 11.84TEE279 pKa = 4.19LGLLLWNTEE288 pKa = 3.99PGAGIATVLAFGSLFAINVLLPALPAYY315 pKa = 8.73WLLVRR320 pKa = 11.84KK321 pKa = 10.19KK322 pKa = 10.26NLRR325 pKa = 3.62
Molecular weight: 36.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1781409 |
32 |
4222 |
379.3 |
42.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.49 ± 0.036 | 0.873 ± 0.024 |
6.122 ± 0.041 | 6.51 ± 0.037 |
4.599 ± 0.023 | 7.911 ± 0.043 |
1.871 ± 0.022 | 5.442 ± 0.026 |
3.803 ± 0.047 | 10.134 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.118 ± 0.019 | 4.183 ± 0.031 |
4.832 ± 0.025 | 3.691 ± 0.023 |
6.102 ± 0.047 | 5.774 ± 0.034 |
5.912 ± 0.05 | 6.722 ± 0.035 |
1.346 ± 0.014 | 3.564 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
