Atta cephalotes (Leafcutter ant)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Hymenoptera; Apocrita; Aculeata; Formicoidea; Formicidae; Myrmicinae; Attini
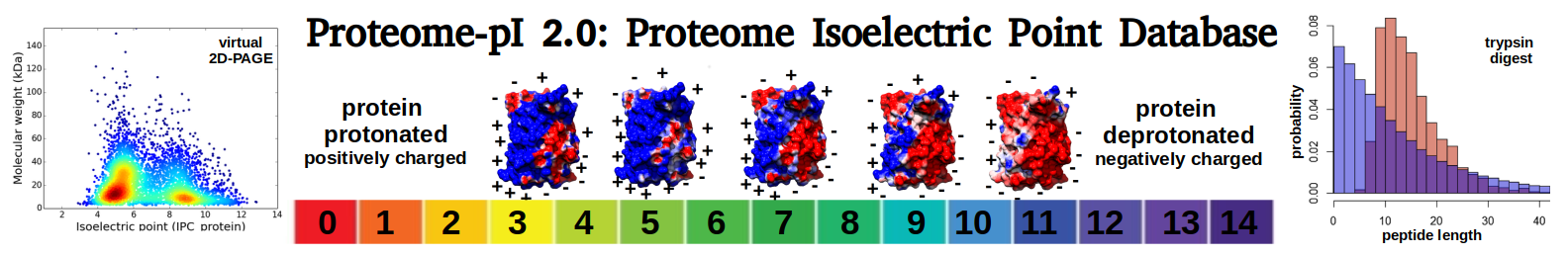
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 10634 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A158NYE1|A0A158NYE1_ATTCE Uncharacterized protein OS=Atta cephalotes OX=12957 GN=105625848 PE=3 SV=1
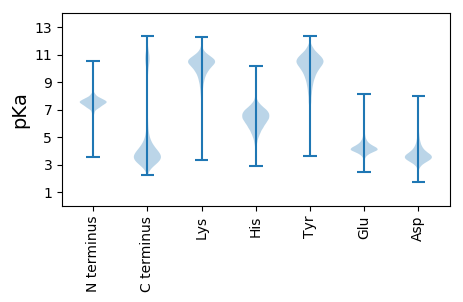
GG1 pKa = 7.27SPLSRR6 pKa = 11.84ADD8 pKa = 3.51LRR10 pKa = 11.84AVNVNLCDD18 pKa = 3.41MCCILSAKK26 pKa = 9.88VPSNDD31 pKa = 4.82DD32 pKa = 2.99PTLADD37 pKa = 4.05KK38 pKa = 10.75EE39 pKa = 4.6AILASLNIKK48 pKa = 10.61AMTFDD53 pKa = 3.55DD54 pKa = 4.76TIGVLSQDD62 pKa = 3.57NEE64 pKa = 4.19XGNLIAVGSPIVLQRR79 pKa = 11.84RR80 pKa = 11.84GSVYY84 pKa = 10.2GANVPMITEE93 pKa = 4.79LVNDD97 pKa = 4.27SNVQFLDD104 pKa = 3.94QDD106 pKa = 4.38DD107 pKa = 5.59DD108 pKa = 4.84DD109 pKa = 6.57DD110 pKa = 6.1PDD112 pKa = 3.87TEE114 pKa = 5.57LYY116 pKa = 8.74LTQPFACGTAFAVSVLDD133 pKa = 3.74SLMSTTYY140 pKa = 10.43FNQNALTLIRR150 pKa = 11.84SLITGGATPEE160 pKa = 4.32LEE162 pKa = 5.17LILAEE167 pKa = 4.32GAGLRR172 pKa = 11.84GGYY175 pKa = 7.88STADD179 pKa = 3.06SLANRR184 pKa = 11.84DD185 pKa = 3.44RR186 pKa = 11.84CRR188 pKa = 11.84VGQIALYY195 pKa = 10.12DD196 pKa = 3.97GPLAQYY202 pKa = 11.29GEE204 pKa = 4.26GGKK207 pKa = 10.74YY208 pKa = 10.55GDD210 pKa = 4.82LFVAALKK217 pKa = 10.61SYY219 pKa = 10.77GMLCIGLYY227 pKa = 10.17
GG1 pKa = 7.27SPLSRR6 pKa = 11.84ADD8 pKa = 3.51LRR10 pKa = 11.84AVNVNLCDD18 pKa = 3.41MCCILSAKK26 pKa = 9.88VPSNDD31 pKa = 4.82DD32 pKa = 2.99PTLADD37 pKa = 4.05KK38 pKa = 10.75EE39 pKa = 4.6AILASLNIKK48 pKa = 10.61AMTFDD53 pKa = 3.55DD54 pKa = 4.76TIGVLSQDD62 pKa = 3.57NEE64 pKa = 4.19XGNLIAVGSPIVLQRR79 pKa = 11.84RR80 pKa = 11.84GSVYY84 pKa = 10.2GANVPMITEE93 pKa = 4.79LVNDD97 pKa = 4.27SNVQFLDD104 pKa = 3.94QDD106 pKa = 4.38DD107 pKa = 5.59DD108 pKa = 4.84DD109 pKa = 6.57DD110 pKa = 6.1PDD112 pKa = 3.87TEE114 pKa = 5.57LYY116 pKa = 8.74LTQPFACGTAFAVSVLDD133 pKa = 3.74SLMSTTYY140 pKa = 10.43FNQNALTLIRR150 pKa = 11.84SLITGGATPEE160 pKa = 4.32LEE162 pKa = 5.17LILAEE167 pKa = 4.32GAGLRR172 pKa = 11.84GGYY175 pKa = 7.88STADD179 pKa = 3.06SLANRR184 pKa = 11.84DD185 pKa = 3.44RR186 pKa = 11.84CRR188 pKa = 11.84VGQIALYY195 pKa = 10.12DD196 pKa = 3.97GPLAQYY202 pKa = 11.29GEE204 pKa = 4.26GGKK207 pKa = 10.74YY208 pKa = 10.55GDD210 pKa = 4.82LFVAALKK217 pKa = 10.61SYY219 pKa = 10.77GMLCIGLYY227 pKa = 10.17
Molecular weight: 24.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A158NRM6|A0A158NRM6_ATTCE Dipeptidase OS=Atta cephalotes OX=12957 GN=105623397 PE=3 SV=1
MM1 pKa = 7.91PPPPPPPSIVVFARR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84MASSSLSSSMPPPLSLPPPPPPPPPPPPPPPPPIGVIVFARR58 pKa = 11.84HH59 pKa = 5.72RR60 pKa = 11.84LASSSSLSSMSSPPSPPNVVVFARR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84LTLSSSSSLL95 pKa = 3.58
MM1 pKa = 7.91PPPPPPPSIVVFARR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84MASSSLSSSMPPPLSLPPPPPPPPPPPPPPPPPIGVIVFARR58 pKa = 11.84HH59 pKa = 5.72RR60 pKa = 11.84LASSSSLSSMSSPPSPPNVVVFARR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84LTLSSSSSLL95 pKa = 3.58
Molecular weight: 9.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
6079146 |
34 |
16023 |
571.7 |
64.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.957 ± 0.023 | 1.984 ± 0.034 |
5.467 ± 0.021 | 6.77 ± 0.041 |
3.579 ± 0.019 | 5.19 ± 0.035 |
2.52 ± 0.013 | 6.315 ± 0.023 |
6.541 ± 0.039 | 9.085 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.404 ± 0.013 | 5.308 ± 0.021 |
4.823 ± 0.028 | 4.357 ± 0.024 |
5.423 ± 0.02 | 8.111 ± 0.033 |
6.034 ± 0.021 | 5.832 ± 0.018 |
1.076 ± 0.007 | 3.21 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
